Page 7 of 10
Re: Mixed Models Module
Posted: Mon Nov 06, 2017 5:17 pm
by Ravi
I'm now looking at the new version and it's looking very nice! One comment about the multinomial model coefficients table. You can group the response groups a bit nicer by using the
combineBelow option for the
estimates table. Basically you just do the following in your
gamljgzlm.r.yaml file:
Code: Select all
- name: dep
title: "Response Groups"
type: text
visible: (modelSelection:multinomial)
combineBelow: true
Another thing that can make the grouping a bit more explicit is by adding some spacing before a new response group begins by using the
addFormat function.
Code: Select all
table$addFormat(rowKey=rowKey, col=1, Cell.BEGIN_GROUP)
So you would only add the format when a row consists of a new response group.
You can see in this screenshot how much of a difference only adding the combineBelow statement already makes:
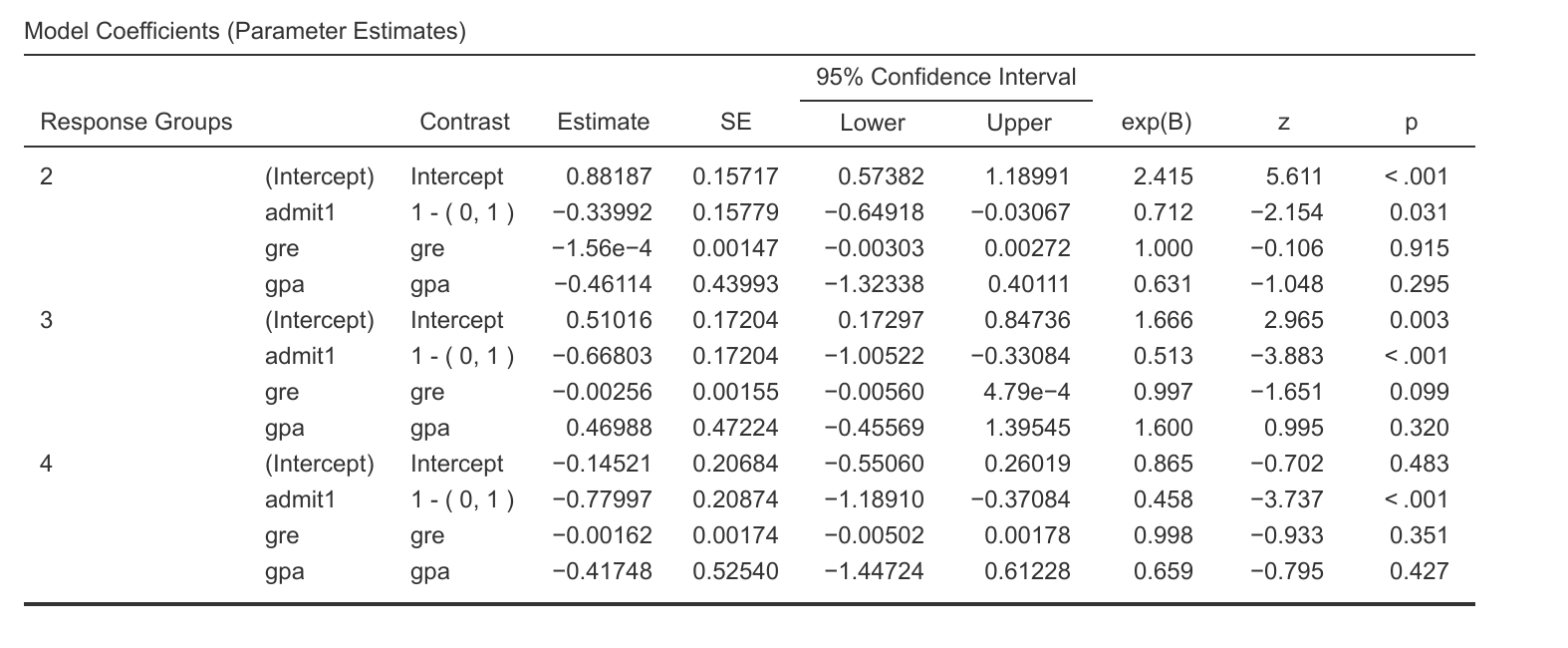
- Screenshot from 2017-11-06 18-16-39.png (166.74 KiB) Viewed 11726 times
Re: Mixed Models Module
Posted: Mon Nov 06, 2017 10:34 pm
by mcfanda@gmail.com
Wow, thanks a lot. I was about to say that I was ready for version 0.0.7 but your suggestions definitely call for 0.0.8. I'm going to implement the "combineBelow" which sounds a very nice option. BTW, I managed to tame nnet::multinom but I should say (WADR) it was a trip in hell. I came out of it with a new motto in R coding : honour thy parent.env() and thy parent.frame()!!!
cheers
mc
Re: Mixed Models Module
Posted: Tue Nov 07, 2017 12:57 pm
by mcfanda@gmail.com
Ok, done. I pushed 0.0.8 with Ravi's suggestions. Tables look better now.
mc
Re: Mixed Models Module
Posted: Tue Nov 07, 2017 3:34 pm
by Ravi
Looks nice! I found a couple of small things

- When doing a multinomial test I get an error when doing post-hoc tests + marginal means:
- In mixed effects model, the contrast column of the parameter estimates table does not update when I change the factor coding. The contrast coding table does. Also, at the moment you have the default factor coding set to deviation. Is it common to use this as default (I would assume simple contrasts would be default).
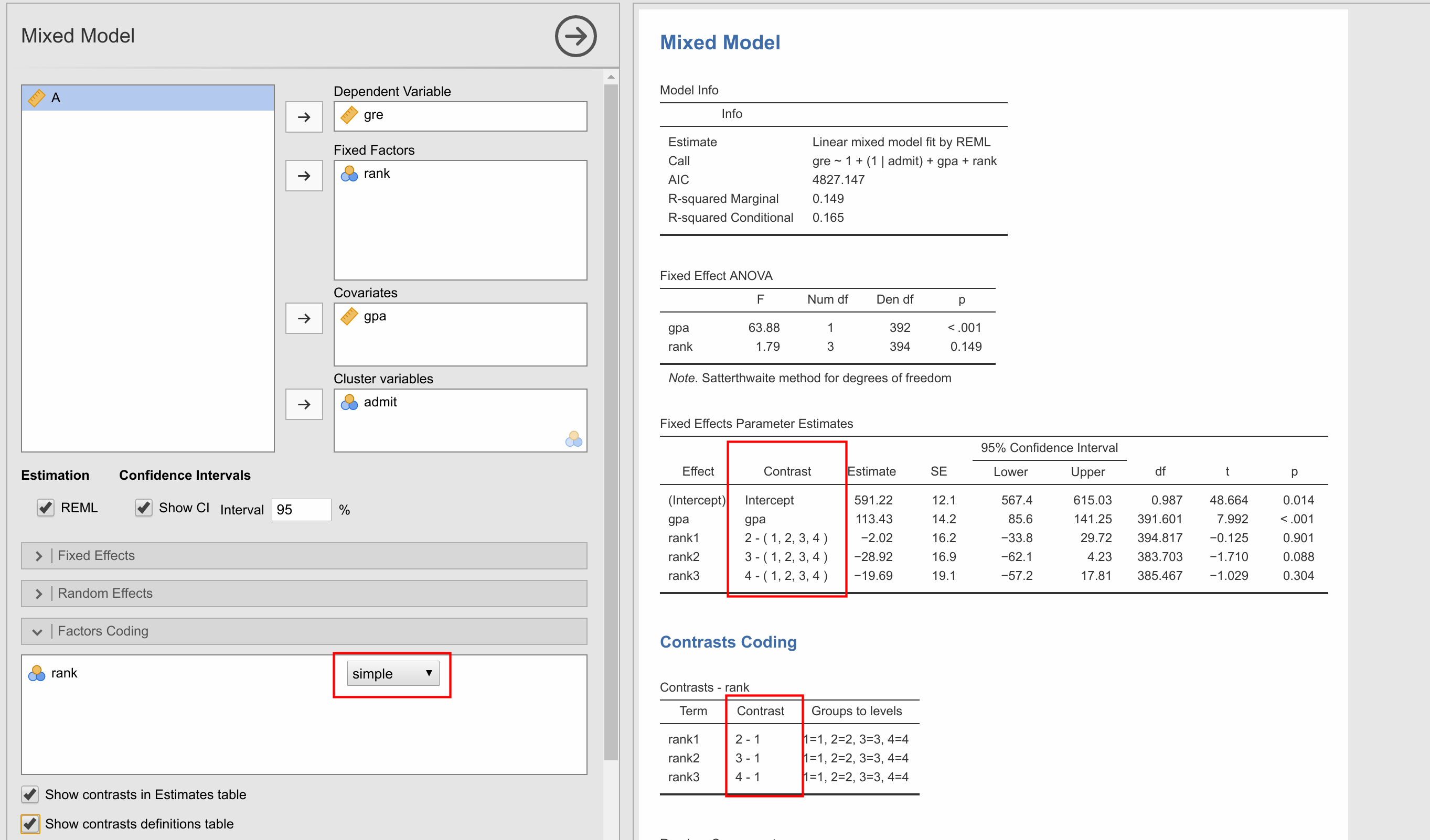
- Screenshot from 2017-11-07 16-18-44.png (276.66 KiB) Viewed 11714 times
- You have changed the results title of GLM to "General Linear Model", but not the options one (in the .a.yaml and .u.yaml). I'm not sure whether this is intentional, but I thought I'd mention it.
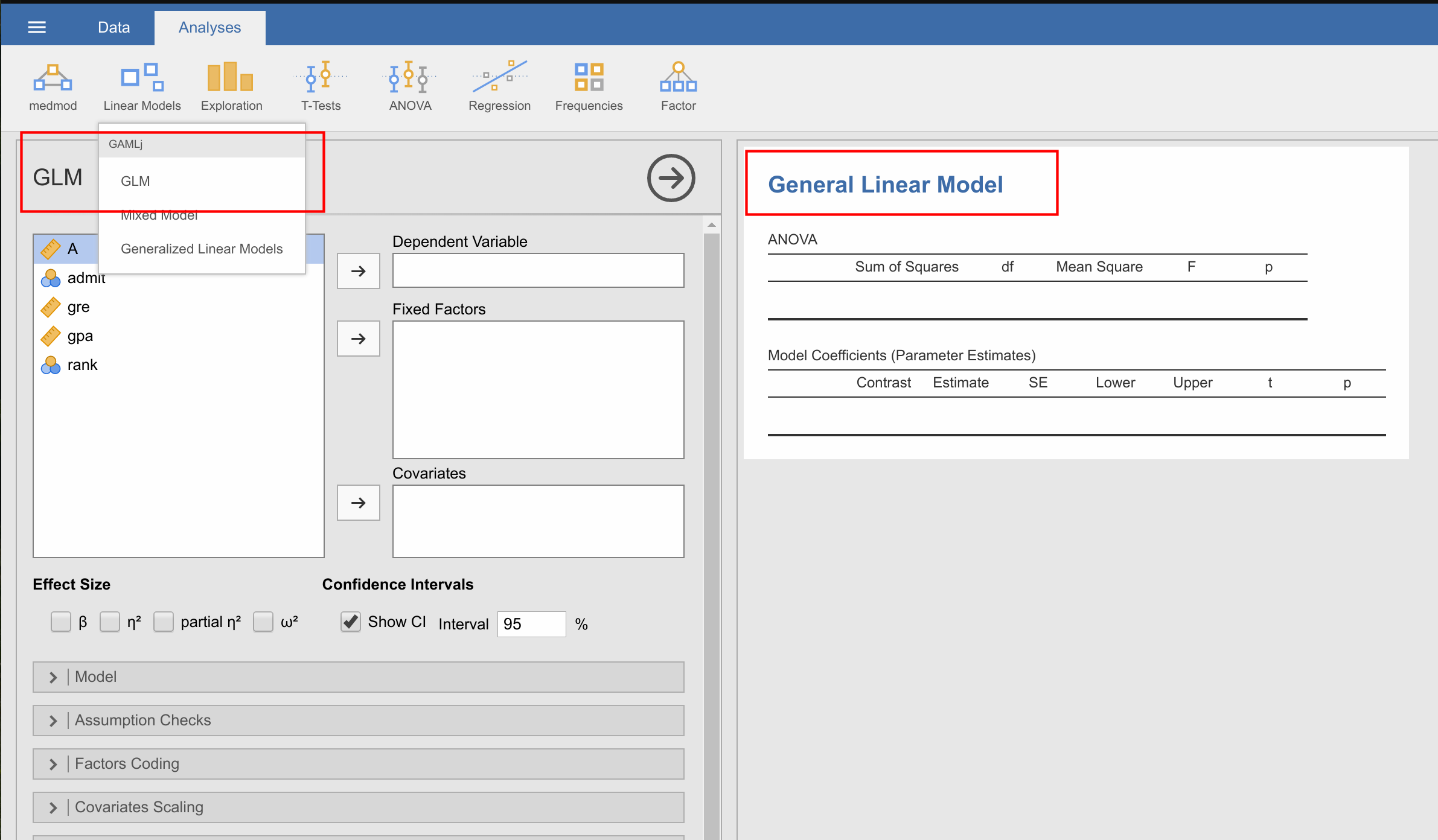
- Screenshot from 2017-11-07 16-22-55.png (150.77 KiB) Viewed 11714 times
But this looks really great and I think it's almost ready for a proper release. How would you like to promote it? Do you have twitter? Of course, we can help you by posting it on our twitter/facebook/linkedin/website, but I think it's good if we can retweet something you have posted so you get the attention you deserve.
Re: Mixed Models Module
Posted: Tue Nov 07, 2017 8:55 pm
by mcfanda@gmail.com
Hi
2-3. fixes implemented (0.0.9)
1. If I load the omv I got an error, but if I save the data in it, open the data and run it , it works fine (in my installation at least) . I probably fixed the bug producing it while updating the rest.
As regard the last point, let me get back to you after I reckon with my lack of ability to promoting things

Re: Mixed Models Module
Posted: Tue Nov 07, 2017 10:39 pm
by jonathon
hi marcello,
i get the same issue as ravi, but perhaps you haven't committed all your changes just yet.
here's the stacktrace:
Code: Select all
Debug
Error in X[ii, ii, drop = FALSE]: (subscript) logical subscript too long
private$.run()
suppressWarnings({
dep <- self$options$dep
factors <- self$options$factors
covs <- self$options$covs
modelType <- self$options$modelSelection
modelTerms <- private$.modelTerms()
if (is.null(dep) || length(modelTerms) == 0)
return()
base::options(contrasts = c("contr.sum", "contr.poly"))
data <- private$.cleanData()
data <- mf.checkData(dep, data, modelType)
if (!is.data.frame(data))
reject(data)
for (factorName in factors) {
lvls <- base::levels(data[[factorName]])
if (length(lvls) == 1)
reject("Factor '{}' contains only a single level", factorName = factorName)
else if (length(lvls) == 0)
reject("Factor '{}' contains no data", factorName = factorName)
}
anovaTable <- self$results$main
estimatesTable <- self$results$estimates
infoTable <- self$results$info
formula <- jmvcore::constructFormula(dep, modelTerms)
formula <- stats::as.formula(formula)
model <- try(private$.estimate(formula, data))
if (isError(model)) {
message <- extractErrorMessage(model)
reject(message)
}
private$.model <- model
self$results$.setModel(model)
infoTable$setRow(rowKey = "r2", list(value = mi.rsquared(model)))
infoTable$setRow(rowKey = "aic", list(value = mf.getAIC(model)))
infoTable$setRow(rowKey = "dev", list(value = model$deviance))
infoTable$setRow(rowKey = "conv", mi.converged(model))
anovaResults <- try(mf.anova(model))
if (isError(anovaResults)) {
message <- extractErrorMessage(anovaResults)
anovaTable$setNote("anocrash", message)
STOP <- TRUE
}
rowNames <- rownames(anovaResults)
for (i in seq_along(rowNames)) {
rowName <- rowNames[i]
tableRow <- anovaResults[i, ]
colnames(tableRow) <- TCONV[["glm.f"]]
anovaTable$setRow(rowNo = i, tableRow)
}
if (mi.aliased(model)) {
infoTable$setRow(rowKey = "conv", list(comm = "Results may be misleading because of aliased coefficients. See Tables notes"))
anovaTable$setNote("aliased", WARNS["ano.aliased"])
estimatesTable$setNote("aliased", WARNS["ano.aliased"])
}
parameters <- try(mf.summary(model))
if (isError(parameters)) {
message <- extractErrorMessage(parameters)
estimatesTable$setNote("sumcrash", message)
STOP <- T
}
ciWidth <- self$options$paramCIWidth/100
if (self$options$showParamsCI) {
citry <- try({
ci <- mf.confint(model, level = ciWidth)
colnames(ci) <- c("cilow", "cihig")
parameters <- cbind(parameters, ci)
})
if (isError(citry)) {
message <- extractErrorMessage(citry)
infoTable$setRow(rowKey = "conv", list(value = "no"))
estimatesTable$setNote("cicrash", paste(message, ". CI cannot be computed"))
}
}
for (i in 1:nrow(parameters)) {
estimatesTable$setRow(rowNo = i, parameters[i, ])
}
if (STOP)
return()
private$.populateSimple(private$.model)
private$.prepareDescPlots(private$.model)
private$.populatePostHoc(model)
private$.populateDescriptives(model)
})
withCallingHandlers(expr, warning = function(w) invokeRestart("muffleWarning"))
private$.populatePostHoc(model)
suppressWarnings({
none <- mf.posthoc(model, ph, "none")
bonferroni <- mf.posthoc(model, ph, "bonferroni")
holm <- mf.posthoc(model, ph, "holm")
})
withCallingHandlers(expr, warning = function(w) invokeRestart("muffleWarning"))
mf.posthoc(model, ph, "none")
.posthoc.multinom(model, ph, "none")
emmeans::emmeans(model, tterm, transform = "response")
ref_grid(object, ...)
regrid(result, transform = transform)
.est.se.df(object, do.se = TRUE)
t(apply(object@linfct[use.elts, , drop = FALSE], 1, function(x) {
if (!any(is.na(x)) && estimability::is.estble(x, object@nbasis, tol)) {
x = x[active]
est = sum(bhat * x)
if (do.se) {
se = sqrt(.qf.non0(object@V, x))
df = object@dffun(x, object@dfargs)
}
else se = df = 0
c(est, se, df)
}
else c(NA, NA, NA)
}))
apply(object@linfct[use.elts, , drop = FALSE], 1, function(x) {
if (!any(is.na(x)) && estimability::is.estble(x, object@nbasis, tol)) {
x = x[active]
est = sum(bhat * x)
if (do.se) {
se = sqrt(.qf.non0(object@V, x))
df = object@dffun(x, object@dfargs)
}
else se = df = 0
c(est, se, df)
}
else c(NA, NA, NA)
})
FUN(newX[, i], ...)
.qf.non0(object@V, x)
Re: Mixed Models Module
Posted: Wed Nov 08, 2017 12:27 am
by MAgojam
Hi Marcello,
I'm looking at the new version (gamlj 0.0.9), if I first use your form, post-hoc error for missing the lsmeans package.
If I do other modules that use it first (post-hoc ANOVA), it also works with your module.
Also reading previous msgs, I've copied lsmeans into gamlj/build/R and recompiled.
Fixed error.
Thanks a lot, you are doing a great job.
Cheers, Maurizio.
Re: Mixed Models Module
Posted: Wed Nov 08, 2017 5:28 am
by jonathon
hey marcello, couple of other things
typos:
generalized linear models => model info => identity not indentity
generalized linear models => 'analysis of deviance' not 'analisys of deviance'
i'd recommend avoiding verbs in the names/titles of options:
i.e. "Contrasts in estimates table", rather than "Show contrasts ..."
"Confidence intervals" rather than "Show CI", etc.
"Show" is implied by the checkbox.
cheers
Re: Mixed Models Module
Posted: Wed Nov 08, 2017 11:32 am
by mcfanda@gmail.com
hey,
@jonathon : fixed and (hopefully) committed and pushed
@maurizio: Yes, you're right. I've moved from lsmeans:: to emmeans: (the new version of the lsmeans package) but I did not clean up all the references. Now it should be ok.
Re: Mixed Models Module
Posted: Thu Nov 09, 2017 5:41 am
by jonathon
looking good! let me know when you want us to push a new version to the library.
you could bump the version number up a bit too if you wanted. something to signal to people that this is more than an alpha.
jonathon